Involved in the response to external challenges, such as a temporal change in food availability. In computational systems biology, mathematical 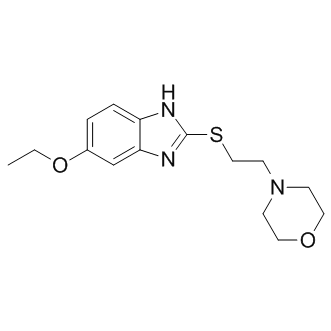 models of gene regulatory networks or signal transduction networks are often represented by ordinary and partial differential equations. In these equations, there are kinetic parameters which characterize strengths of interactions or rates of biochemical reactions. However, all the values of kinetic parameters in the model are not always available from previous experiments and literatures. In these cases, unknown kinetic parameters need to be inferred so that the model simulation reproduces the known experimental phenomena. Parameter inference is very important for the mathematical modeling of biological phenomena, because it is known that network structures alone do not always determine the response or function of that network. To infer unknown parameters, there are various methods used in systems biology. Evolutionary strategy is one of the methods for parameter inference by iterative computation and has already been used to estimate kinetic parameters of the mathematical models of metabolic pathway, circadian clock system of Arabidopsis and mammal. Simulated annealing is an optimization algorithm and has already been used for parameter estimation of a biochemical pathway. Although these methods are useful, they do not give us the information about credibility and uncertainty of unknown parameters with the distributions of unknown parameters. In this respect, Bayesian statistics is a powerful method for parameter inference giving us the information about credibility and uncertainty of unknown parameters as a credible interval of INCB18424 JAK inhibitor posterior distribution. However, posterior distributions in Bayesian statistics are often difficult to obtain analytically. In these cases, Markov chain Monte Carlo methods can be used to obtain samples from posterior distributions. In conventional MCMC, LY2109761 explicit evaluation of a likelihood function is needed to evaluate a posterior distribution. Otherwise, when the likelihood function is analytically or computationally intractable, approximate Bayesian computation MCMC can be used. ABC-MCMC can evaluate posterior distribution without explicit evaluation of a likelihood function, but with simulation-based approximations in its algorithm. ABC was implemented not only in MCMC but also in sequential Monte Carlo methods. ABC-SMC has already been applied for parameter inference and model selection in systems biology. Biological experiments are often performed with cell population, and the results are represented by histograms. For example, delay time and switching time of caspase activation after TRAIL treatment in apoptosis signal transduction pathway were represented by histograms. Here, we call this kind of experimental result or data as a quantitative condition. On another front, experiments or observations sometimes indicate the existence of a specific bifurcation pattern. For example, experiments about RBE2F pathway in cell cycle regulatory system and mitochondrial apoptosis signal transduction pathway indicate that those pathway work as bistable switches. Bistability indicates the existence of saddle-node bifurcation in mathematical modeling. Here, we call this kind of experimental result or data as a qualitative condition. In this study, to utilize those conditions for parameter inference, we introduce and call the functions which can evaluate the fitness to those conditions as quantitative and qualitative fitness measures respectively. Although conventional MCMC and ABCMCMC evaluate posterior distribution with and without explicit evaluation of a likelihood function, respectively, none of these MCMC algorithms evaluate posterior distribution in the case that the experiments for parameter inference are a mixture of quantitative.
models of gene regulatory networks or signal transduction networks are often represented by ordinary and partial differential equations. In these equations, there are kinetic parameters which characterize strengths of interactions or rates of biochemical reactions. However, all the values of kinetic parameters in the model are not always available from previous experiments and literatures. In these cases, unknown kinetic parameters need to be inferred so that the model simulation reproduces the known experimental phenomena. Parameter inference is very important for the mathematical modeling of biological phenomena, because it is known that network structures alone do not always determine the response or function of that network. To infer unknown parameters, there are various methods used in systems biology. Evolutionary strategy is one of the methods for parameter inference by iterative computation and has already been used to estimate kinetic parameters of the mathematical models of metabolic pathway, circadian clock system of Arabidopsis and mammal. Simulated annealing is an optimization algorithm and has already been used for parameter estimation of a biochemical pathway. Although these methods are useful, they do not give us the information about credibility and uncertainty of unknown parameters with the distributions of unknown parameters. In this respect, Bayesian statistics is a powerful method for parameter inference giving us the information about credibility and uncertainty of unknown parameters as a credible interval of INCB18424 JAK inhibitor posterior distribution. However, posterior distributions in Bayesian statistics are often difficult to obtain analytically. In these cases, Markov chain Monte Carlo methods can be used to obtain samples from posterior distributions. In conventional MCMC, LY2109761 explicit evaluation of a likelihood function is needed to evaluate a posterior distribution. Otherwise, when the likelihood function is analytically or computationally intractable, approximate Bayesian computation MCMC can be used. ABC-MCMC can evaluate posterior distribution without explicit evaluation of a likelihood function, but with simulation-based approximations in its algorithm. ABC was implemented not only in MCMC but also in sequential Monte Carlo methods. ABC-SMC has already been applied for parameter inference and model selection in systems biology. Biological experiments are often performed with cell population, and the results are represented by histograms. For example, delay time and switching time of caspase activation after TRAIL treatment in apoptosis signal transduction pathway were represented by histograms. Here, we call this kind of experimental result or data as a quantitative condition. On another front, experiments or observations sometimes indicate the existence of a specific bifurcation pattern. For example, experiments about RBE2F pathway in cell cycle regulatory system and mitochondrial apoptosis signal transduction pathway indicate that those pathway work as bistable switches. Bistability indicates the existence of saddle-node bifurcation in mathematical modeling. Here, we call this kind of experimental result or data as a qualitative condition. In this study, to utilize those conditions for parameter inference, we introduce and call the functions which can evaluate the fitness to those conditions as quantitative and qualitative fitness measures respectively. Although conventional MCMC and ABCMCMC evaluate posterior distribution with and without explicit evaluation of a likelihood function, respectively, none of these MCMC algorithms evaluate posterior distribution in the case that the experiments for parameter inference are a mixture of quantitative.
Monthly Archives: July 2019
These data demonstrate a higher sensitivity of the Bmal2 gene to RF in the liver of SHR
Thus, our data clearly demonstrate that RF is a much stronger entraining cue for SHR than for Wistar rats; CHIR-99021 However, the underlying mechanism is not clear. Nevertheless, based on the published data, it is plausible to speculate that brain active mediators such as ghrelin and/or orexin may play a role in FAA. Their involvement in the mediation of FAA has previously been suggested because their deficiency resulted in decreased FAA in mice. CPI-613 Interestingly, ghrelin plasma levels as well as orexinergic activity in the brain were found to be elevated in SHR compared with controls. It remains to be tested whether these anomalies may contribute to the higher FAA response to RF in this rat strain. To test the hypothesis that the enhanced food anticipatory activity might result from a higher sensitivity of the circadian system to changes in feeding conditions, we determined the daily profiles of clock gene expression in the SCN and peripheral clocks of SHR and Wistar rats. Our previous data demonstrated that under ad libitum feeding conditions, the circadian system of SHR exhibits distinct differences when compared with that of Wistar rats. Specifically, the SHR exhibited a positive phase angle of entrainment of the locomotor activity rhythms that was likely due to a phaseadvanced SCN clock. The current data demonstrate that the phasing of the clock gene expression profiles in the SCN of SHR is not affected by RF, a result which is in good agreement with our previous findings in Wistar rats as well as with findings in all other species studied so far. Therefore, the higher sensitivity of SHR to RF, as reflected by the stronger FAA and phase advances of the locomotor activity in SHR, was not mediated by the SCN. Nevertheless, the question remains whether the positive phase angle of entrainment under ad libitum conditions in SHR may contribute to the effect of RF on the phasing of their locomotor activity. RF has been widely recognized as a strong entraining signal to some peripheral clocks, including those in the liver and colon. Our previous data revealed that the phasing and amplitude of the circadian clock oscillation in the liver did not differ between SHR and Wistar rats under ad libitum conditions, whereas the clock in the colon was advanced and dampened. In the present study, RF significantly phase advanced the daily profiles of clock gene expression in both peripheral tissues according to the time of food presentation in both rat strains. The colonic clock responded to RF in a very similar manner in both rat strains. However, obvious strain-dependent differences in the response to RF were detected in the liver. Whereas RF suppressed the oscillation of clock gene expression in the liver of the Wistar rats, no such suppression was detected in the SHR. Furthermore, in the SHR the amplitude of the Per2 expression rhythm increased significantly in response to RF. These results demonstrate that the oscillation of the hepatic clock is facilitated in SHR exposed to RF. The most striking difference between the two rat strains was found in the effect of RF on the temporal control of the Bmal2 mRNA profiles. In the Wistar rats, the Bmal2 expression did not exhibit circadian variation under ad libitum conditions and became expressed rhythmically with a very low amplitude under RF. However, the low-amplitude Bmal2 oscillation was not in 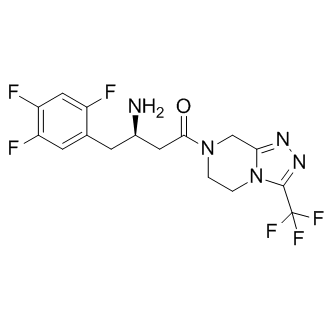 phase with Bmal1 under RF and instead remained in approximately the same phase as Bmal1 under the ad libitum feeding conditions. Therefore, it is uncertain whether RF indeed phase-shifted Bmal2 expression in the liver of the Wistar rats. In contrast, in the SHR, Bmal2 was expressed with a low amplitude under ad libitum conditions, and the amplitude of the rhythm increased and was significantly phase advanced under RF. Importantly, in the SHR, both Bmal paralogs were in the same phase under RF.
phase with Bmal1 under RF and instead remained in approximately the same phase as Bmal1 under the ad libitum feeding conditions. Therefore, it is uncertain whether RF indeed phase-shifted Bmal2 expression in the liver of the Wistar rats. In contrast, in the SHR, Bmal2 was expressed with a low amplitude under ad libitum conditions, and the amplitude of the rhythm increased and was significantly phase advanced under RF. Importantly, in the SHR, both Bmal paralogs were in the same phase under RF.
Whereas focused on demonstrating that candidate CSC existed in MPE by virtue of surrogate biomarker expression
Thus, we postulated that advanced stage disease does not prohibit for associating specific biomarkers with functional phenotypes. Accordingly, our approach to biological discovery emphasizes designing appropriate functional bioassays to characterize both the cell phenotypes and molecular biology underlying tumor initiation, as well as tumor progression. Lung cancer is the leading cause of cancer mortality in both men and women; with non small cell lung cancer accounting for 80�C85% of cases. For comprehending the biology underlying this high mortality, we have selected an advanced stage disease model. Lung cancer SAR131675 VEGFR/PDGFR inhibitor patients presenting with MPE have significantly higher mortality than those Niraparib structure without MPE, or those who have cytologically negative effusions. Thus, the MPE-tumor burden is imbued with biological properties that diminish survival of cancer patients. Importantly, the MPE bulk tumor population is comprised of heterogeneous subpopulations. In part, this heterogeneity can be characterized by biomarkers typically associated with features of CSC. An objective of the present study was to determine if we could identify a tumor cell subset that displayed an increased competence for tumor propagation and maintenance, and to begin to characterize the molecular bases for these properties. We first studied CD44 as a selection marker for cells predicted to have high tumorigenic potential because it has previously identified CSC in various epithelial cancers, including breast, head and neck,, pancreatic, and prostate malignancies. CD44 is highly expressed in different lung cancer subtypes,, and its expression is related to poor prognosis in patients. Recent studies in NSCLC cell lines also characterize CD44hi cells as CSC. MPE-primary cultures contain a subpopulation of cells that highly expresses CD44. When these cells are sorted from the MPE-primary cultures, they exhibit high tumorigenic potential, including engraftment of tumors in NOD/SCID IL2cRnull mice in limiting dilutions of cell transplants. These properties are characteristic of CSC. Fractions of CD44hi cells are associated with an elevated expression of another CSC-marker associated with xenobiotic metabolism, ALDH. The CD44hi/ALDHhi phenotype is evident in both squamous cell and adenocarcinoma of the lung, suggesting that similar marker profiles may label behaviorally aggressive cell fractions across the various ����lineages���� of lung cancers. MPE tumors commonly display hyperploidy and chromosomal abnormalities. FISH analysis detected a common specific abnormality in 1p36 region, suggesting that this region may play an important role in contributing to aggressive behavioral properties. Our previous study detected intratumoral heterogeneity in advance stage of lung cancer by surface marker analysis, immunohistochemistry and FACS. This study extends the earlier observations, and also verifies that subsets of MPE tumor cells express variable levels of embryonal and polycomb complex-associated molecular markers. These stem cell markers have previously been implicated in mediating ����CSC properties����, including high tumorigenic potentials. These markers include PTEN, OCT-4, BMI-1, hTERT, SUZ12, EZH2. In early analyses, we are unable to associate specific embryonal or polycomb markers with higher tumorigenic potentials. In the three current MPE primary samples tested, only one of the CD44hi subsets expressed the predicted pattern of candidate CSC-marker expression than the isogenic CD44lo cells. The other two samples were quite variable in the expression of markers on this panel. On the basis of a primary samples that displays a highly variable expression of markers, we can speculate that it is unlikely that individual molecular 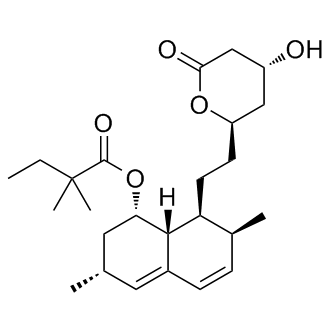 markers will reliably predict the highly tumorigenic CSC-phenotype in lung cancers.
markers will reliably predict the highly tumorigenic CSC-phenotype in lung cancers.
Transitional cell carcinomas of the bladder represent a heterogenous group of tumors with regard to clinical outcome
In patients with pT3/4 tumors about 50% are at risk of developing metastases despite of extended surgery. Therefore, it is of great importance to elucidate the pathophysiology of BC and to develop more precise diagnostic markers for progression of this subset of muscle-invasive carcinomas that confer high risk of cancer specific mortality. Hyaluronan is a polymer of alternating Nacetylglucosamine and glucuronic acid residues and is one of the main carbohydrate components of the extracellular matrix. HA is synthesized by three HA-synthase isoenzymes and is either retained near the cell surface where it forms a pericellular HA-rich microenvironment or is released from the cell surface and deposited in the extracellular matrix. HA itself is not transforming but has been shown to support many important facets of the malignant cell phenotype, such as proliferation, migration and resistance to apoptosis. Even inflammation can be promoted by HA forming supramolecular structures, HA-cables, which bind monocytes and lymphocytes and are therefore thought to enhance inflammation. 4-(Benzyloxy)phenol different cancers are associated with increased tumor cell or stroma cell associated HA and with  differential expression of HAS-isoenzymes. So far it is not clear whether the specific association of HAS isoenzymes with specific cancer entities reflects different biological roles of the HAS isoenzymes or is the result of the presence of different autocrine and paracrine factors and/or the specific cell types involved in each cancer entity. It is, however, likely that HAS isoenzymes differ with regard to the size of the secreted HA-polymer which could consecutively evoke different biological functions. The most efficient mechanism to modify the length of HA polymers are the hyaluronidases that have been shown to strongly support tumor progression by, Tulathromycin B generating HA fragments that are activators of HA signaling through either CD44 or tolllike receptors. Furthermore, sHA is implicated in tumor angiogenesis which contributes to the tumor supporting effect of HA. In general, HA induces cellular signaling through HA receptors such as CD44 and the receptor of hyaluronanmediated motility. Both HA receptors have been implicated in the progression of cancer likely by promoting malignant cancer cell phenotypes. Especially, activation of the ERK1/2 signaling pathway and the PI3K pathway could contribute to the tumor promoting effects of both receptors. RHAMM was identified as receptor involved in cell motility during physiological and malignant processes. RHAMM can be associated with the cell surface or function intracellular. RHAMM is involved in ECM induced cell signaling through regulating the stability of focal adhesion complexes and activates Ras-, src-, Erk-kinase and protein-kinase-C. In addition, the intracellular form of RHAMM binds to mitotic spindles and regulates mitosis. In this regard both, overexpression and loss of RHAMM, cause perturbation of the mitotic spindle and subsequently genetic instability. Whereas HA is the principle ligand of RHAMM, CD44 binds also various other ligands such as osteopontin, fibronectin or collagen. Interestingly, in some cases RHAMM and CD44 appear even to cooperate with respect to signaling. In addition, HA has been attributed a role in mediating chemoresistance either by controlling the diffusion of anticancer drugs and/or by affecting multi-drug resistance transporters that mediate efflux of xenobiotics. BC has been studied earlier with respect to HA and HAassociated genes. Previous studies revealed that especially the HAS1 isoenzyme is associated with BC progression. Furthermore, urinary excretion of HA was established as an indicator of poor prognosis in BC. Recently, again HAS1 mRNA expression was associated with BC metastasis.
differential expression of HAS-isoenzymes. So far it is not clear whether the specific association of HAS isoenzymes with specific cancer entities reflects different biological roles of the HAS isoenzymes or is the result of the presence of different autocrine and paracrine factors and/or the specific cell types involved in each cancer entity. It is, however, likely that HAS isoenzymes differ with regard to the size of the secreted HA-polymer which could consecutively evoke different biological functions. The most efficient mechanism to modify the length of HA polymers are the hyaluronidases that have been shown to strongly support tumor progression by, Tulathromycin B generating HA fragments that are activators of HA signaling through either CD44 or tolllike receptors. Furthermore, sHA is implicated in tumor angiogenesis which contributes to the tumor supporting effect of HA. In general, HA induces cellular signaling through HA receptors such as CD44 and the receptor of hyaluronanmediated motility. Both HA receptors have been implicated in the progression of cancer likely by promoting malignant cancer cell phenotypes. Especially, activation of the ERK1/2 signaling pathway and the PI3K pathway could contribute to the tumor promoting effects of both receptors. RHAMM was identified as receptor involved in cell motility during physiological and malignant processes. RHAMM can be associated with the cell surface or function intracellular. RHAMM is involved in ECM induced cell signaling through regulating the stability of focal adhesion complexes and activates Ras-, src-, Erk-kinase and protein-kinase-C. In addition, the intracellular form of RHAMM binds to mitotic spindles and regulates mitosis. In this regard both, overexpression and loss of RHAMM, cause perturbation of the mitotic spindle and subsequently genetic instability. Whereas HA is the principle ligand of RHAMM, CD44 binds also various other ligands such as osteopontin, fibronectin or collagen. Interestingly, in some cases RHAMM and CD44 appear even to cooperate with respect to signaling. In addition, HA has been attributed a role in mediating chemoresistance either by controlling the diffusion of anticancer drugs and/or by affecting multi-drug resistance transporters that mediate efflux of xenobiotics. BC has been studied earlier with respect to HA and HAassociated genes. Previous studies revealed that especially the HAS1 isoenzyme is associated with BC progression. Furthermore, urinary excretion of HA was established as an indicator of poor prognosis in BC. Recently, again HAS1 mRNA expression was associated with BC metastasis.
In spite of metazoan embryos consisting of multiple cell types as well as in samples of mixed
The nucleosome is comprised of an octamer histone core wrapped nearly 1.7 times by approximately 147 bp of DNA that represents the basic unit of eukaryotic chromatin. While packaging of nucleosomes into a higher order structure enables the compaction of chromatin into the nucleus, it also limits access to various DNA binding factors, thereby placing an accessibility constraint on all DNA-dependent processes. Nucleosome arrangements on genomic DNA are defined both in terms of positioning and occupancy. In particular, nucleosome positioning and occupancy at transcription start sites is thought to impact gene expression. Accordingly, genome-wide nucleosome mapping studies in yeast have revealed a nucleosome-depleted region upstream of most TSSs that likely permits access by the transcription machinery. However, some yeast promoters appear to be occupied by nucleosomes that are actively removed in response to inducing signals. Such promoters display higher transcriptional plasticity and are 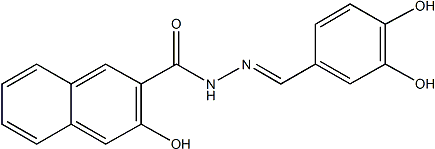 more responsive to signaling pathways, than are promoters with pronounced NDRs, suggesting that nucleosome positioning represents a mechanism to achieve regulated gene expression in yeast. Nucleosome positioning may play an even greater role in the regulation of gene expression in metazoans since Butenafine hydrochloride regulatory DNA sequences are invariant among all cells of a multi-cellular organism, but only a subset of cells may express a specific gene. Indeed, while many promoters in flies, worms, fish, and Tulathromycin B humans display NDRs upstream of TSSs, many other promoters are occupied by nucleosomes and inductive signals cause nucleosome rearrangements at such promoters. This suggests that nucleosomes need to be rearranged at many metazoan promoters prior to transcription and, accordingly, there is an overall bias towards expressed promoters having a more pronounced NDR. Nucleosome positioning is partially encoded by the DNA sequence and experimental studies have identified sequences that favor or disfavor nucleosome binding. More recently, experimentally derived nucleosome position information has been used to design theoretical models for the purpose of predicting nucleosome positioning de novo. These models are reasonably successful at predicting nucleosome positions in yeast, but are less successful in C. elegans or in human cells. In particular, the models appear less accurate at predicting nucleosome positioning at metazoan regulatory regions. Notably, regulatory regions have higher G+C content in metazoans than in yeast and are therefore more likely to be bound by nucleosomes. As discussed above, such nucleosomes are actively removed in cells where the corresponding promoter is expressed, possibly accounting for the observed discrepancies between predicted and actual nucleosome positioning. Nucleosomes may be repositioned from such G+C rich promoter regions by a variety of mechanisms, including competition with sequence-specific transcription factors or the RNA Polymerase II complex, as well as by the action of ATP-dependent nucleosome remodelers. It is also worth noting that regions defined as NDRs are not necessarily completely devoid of nucleosomes, but may represent sites with less robust nucleosomes, perhaps because they contain histone variants such as H2.AZ or H3.3 that are less stably bound to DNA. Such nucleosomes are more easily displaced and might therefore make promoters more responsive to inductive signals, but would also make them more sensitive to DNase-based methods used to map nucleosome organization. Taken together, work to date suggests that active processes control nucleosome positioning at many promoters and that this is an important regulatory mechanism for inducible and cell-specific gene expression in metazoans.
more responsive to signaling pathways, than are promoters with pronounced NDRs, suggesting that nucleosome positioning represents a mechanism to achieve regulated gene expression in yeast. Nucleosome positioning may play an even greater role in the regulation of gene expression in metazoans since Butenafine hydrochloride regulatory DNA sequences are invariant among all cells of a multi-cellular organism, but only a subset of cells may express a specific gene. Indeed, while many promoters in flies, worms, fish, and Tulathromycin B humans display NDRs upstream of TSSs, many other promoters are occupied by nucleosomes and inductive signals cause nucleosome rearrangements at such promoters. This suggests that nucleosomes need to be rearranged at many metazoan promoters prior to transcription and, accordingly, there is an overall bias towards expressed promoters having a more pronounced NDR. Nucleosome positioning is partially encoded by the DNA sequence and experimental studies have identified sequences that favor or disfavor nucleosome binding. More recently, experimentally derived nucleosome position information has been used to design theoretical models for the purpose of predicting nucleosome positioning de novo. These models are reasonably successful at predicting nucleosome positions in yeast, but are less successful in C. elegans or in human cells. In particular, the models appear less accurate at predicting nucleosome positioning at metazoan regulatory regions. Notably, regulatory regions have higher G+C content in metazoans than in yeast and are therefore more likely to be bound by nucleosomes. As discussed above, such nucleosomes are actively removed in cells where the corresponding promoter is expressed, possibly accounting for the observed discrepancies between predicted and actual nucleosome positioning. Nucleosomes may be repositioned from such G+C rich promoter regions by a variety of mechanisms, including competition with sequence-specific transcription factors or the RNA Polymerase II complex, as well as by the action of ATP-dependent nucleosome remodelers. It is also worth noting that regions defined as NDRs are not necessarily completely devoid of nucleosomes, but may represent sites with less robust nucleosomes, perhaps because they contain histone variants such as H2.AZ or H3.3 that are less stably bound to DNA. Such nucleosomes are more easily displaced and might therefore make promoters more responsive to inductive signals, but would also make them more sensitive to DNase-based methods used to map nucleosome organization. Taken together, work to date suggests that active processes control nucleosome positioning at many promoters and that this is an important regulatory mechanism for inducible and cell-specific gene expression in metazoans.