Previous work from our laboratory has demonstrated that TGinduced apoptosis as well as apoptosis resistance induced by EGF in DU145 cells are mechanisms dependent of intracellular Ca2+ signalling. We have observed that PHE treatment has no affect on the intracellular Ca2+ concentration. This observation strongly suggests that resistance to TG-induced apoptosis in DU145 by PHE does not involve the Ca2+ signalling pathway. a1A-AR signalling is complex and various lines of evidence demonstrate that a1A-AR stimulation can activate a variety of signalling pathways depending on cell type, receptor expression and agonist concentration. It has been previously described, in cardiomyocytes, that a1A-AR activate ERK-related survival pathways. We have revealed, for the first time, the involvement of the ERK1/2 MAPK pathway in the apoptosis resistance function of the a1A-AR in DU145 androgenindependent prostate cancer cells. Interestingly, it has been demonstrated that human prostate biopsies have increased 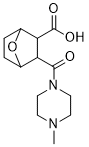 levels of activated ERKs in malignant tissues as compared to those in benign specimens. In conclusion, we demonstrate that caveolae of androgenindependent prostate cancer cells DU145 are involved in the apoptosis resistance induced by the a1A-AR. Our data show that caveolae integrity is necessary for a1A-AR function in these cells. This study reveals that a1A-AR and cav-1 expressions are positively correlated with the pathological state of human prostate and are highly expressed in advance PCa tissue samples. Our data strongly suggest that the protective function of the a1A-AR via caveolae described in DU145 cells, may contribute in synergy with other factors, to the apoptosis resistance mechanisms of PCa cells and enhancement of their survival. Aggregation of hyper-phosphorylated protein tau into filaments and eventually neurofibrillary tangles is characteristic for tauopathies, a large and diverse group of neurodegenerative Tulathromycin B disorders, including Alzheimer��s disease. Primary tauopathies present as clinically variable entities, e.g. Pick��s disease, progressive supranuclear palsy, corticobasal degeneration and frontotemporal dementia, among others. Tauopathy is defined by fibrillar and tangled aggregates of phosphorylated protein tau, which is normally a very soluble protein that binds to microtubules to secure their assembly, stability and spacing. Tau3R and Tau4R isoforms have different affinity for microtubules and their relative abundance is regulated by alternative mRNA splicing. Post-translational, dynamic regulation of microtubule-binding is thought to occur by phosphorylation of tau at various serine/threonine residues by various kinases, including GSK3, cdk5, and MARK, among others. In adult ageing brain, in primary tauopathies and in AD, protein tau becomes excessively phosphorylated, eventually changing its conformation to induce aggregation resulting in tauopathy. Interestingly, both intronic and exonic mutations in the MAPT gene encoding protein tau, are dominantly associated with various tauopathies implying that neurotoxicity results from mutant tau protein, but as well from wildtype tau by 4-(Benzyloxy)phenol isoform imbalances. Alzheimer��s disease is the most prominent secondary tauopathy, wherein intracellular tau inclusions combine with amyloid deposits. Amyloid peptides are normal constituents in human brain at any age, stemming from amyloid precursor protein by a complex set of proteinases. With ageing, amyloid peptides accumulate and aggregate, eventually becoming deposited in parenchym and vasculature, even in cognitive normal individuals as is emerging from clinical imaging studies.
levels of activated ERKs in malignant tissues as compared to those in benign specimens. In conclusion, we demonstrate that caveolae of androgenindependent prostate cancer cells DU145 are involved in the apoptosis resistance induced by the a1A-AR. Our data show that caveolae integrity is necessary for a1A-AR function in these cells. This study reveals that a1A-AR and cav-1 expressions are positively correlated with the pathological state of human prostate and are highly expressed in advance PCa tissue samples. Our data strongly suggest that the protective function of the a1A-AR via caveolae described in DU145 cells, may contribute in synergy with other factors, to the apoptosis resistance mechanisms of PCa cells and enhancement of their survival. Aggregation of hyper-phosphorylated protein tau into filaments and eventually neurofibrillary tangles is characteristic for tauopathies, a large and diverse group of neurodegenerative Tulathromycin B disorders, including Alzheimer��s disease. Primary tauopathies present as clinically variable entities, e.g. Pick��s disease, progressive supranuclear palsy, corticobasal degeneration and frontotemporal dementia, among others. Tauopathy is defined by fibrillar and tangled aggregates of phosphorylated protein tau, which is normally a very soluble protein that binds to microtubules to secure their assembly, stability and spacing. Tau3R and Tau4R isoforms have different affinity for microtubules and their relative abundance is regulated by alternative mRNA splicing. Post-translational, dynamic regulation of microtubule-binding is thought to occur by phosphorylation of tau at various serine/threonine residues by various kinases, including GSK3, cdk5, and MARK, among others. In adult ageing brain, in primary tauopathies and in AD, protein tau becomes excessively phosphorylated, eventually changing its conformation to induce aggregation resulting in tauopathy. Interestingly, both intronic and exonic mutations in the MAPT gene encoding protein tau, are dominantly associated with various tauopathies implying that neurotoxicity results from mutant tau protein, but as well from wildtype tau by 4-(Benzyloxy)phenol isoform imbalances. Alzheimer��s disease is the most prominent secondary tauopathy, wherein intracellular tau inclusions combine with amyloid deposits. Amyloid peptides are normal constituents in human brain at any age, stemming from amyloid precursor protein by a complex set of proteinases. With ageing, amyloid peptides accumulate and aggregate, eventually becoming deposited in parenchym and vasculature, even in cognitive normal individuals as is emerging from clinical imaging studies.
Monthly Archives: May 2019
Why AAV-based models are more powerful in producing neuro-degeneration than transgenic models
Hippocampal neurons in our mice that model hippocampal sclerosis by inducible expression of p25, the truncated activator of cdk5. Other pathological features like tangles, spheroids and axonal dilatations were revealed by various staining methods, but only sporadically and not consistently in all AAV-Tau injected mice. Neither their localization nor distribution nor abundance could explain the observed massive pyramidal cell-death. This remarkable outcome corroborates our previous observation in bigenic mice with massive forebrain tauopathy, i.e. fibrillar tau is not neurotoxic per se. Moreover, the combined data support the hypothesis that fibrillar tau-aggregates function as a sink, to protect neurons against smaller tauaggregates or -oligomers that are the real neurotoxic species. The biochemical and physical properties of the postulated neurotoxic tau-oligomers can eventually be defined in the AAVmodel, and compared to those in Catharanthine sulfate transgenic models. Nevertheless, that might prove a non-conclusive exercise, as discussed below. Particularly mice with inducible tau.P301L expression showed neurodegeneration and tangles but at much higher expression levels of the mutant tau.P301L. To our knowledge, models based on wild-type Tau.4R have not been reported to suffer appreciable neurodegeneration in the hippocampus. We analyzed the phosphorylation status of tau and collected extensive biochemical and immuno-histochemical data-sets on tau phospho-epitopes in the AAV models, as in our transgenic mice. No specific single or complex phospho-epitope on tau is identified as essential for, or directly related to the neurodegeneration. This is not unexpected, and in line with many studies on human brain and in experimental models, supporting the conviction that variable Mepiroxol combinations of phosphorylations of protein tau underlie its neurotoxicity. By logical extension, not a single kinase but combined actions of several kinases are to be hold responsible for phosphorylating tau to make it eventually harmful to neurons. Moreover, the sets of phosphoepitopes and of kinases most likely will vary pending the affected brain region, i.e. the disorder. Because tauopathies vary widely in their clinical, pathological and biochemical characteristics, the molecular identification of a unique, unifying neurotoxic tauspecies becomes more and more unlikely. We first and foremost consider important the only known physiological function of protein tau, i.e. binding to microtubules. If phosphorylated protein tau fails to stabilize microtubules, or affects microtubule dynamics, a dysfunctional cytoskeleton with axonal, dendritic and synaptic defects as results. The observed changes in cytoarchitecture in degenerating neurons, support the hypothesis that they contribute to the overall process. The experimental data obtained with truncated Tau255 most strongly imply that microtubule binding of protein tau is essential in inflicting neurodegeneration and 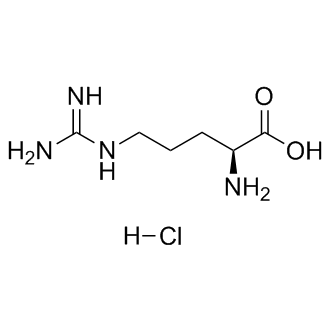 involve the microtubular network as a structural and transport scaffold. The alternative explanation, i.e. that wild-type and mutant full-length Tau but not truncated Tau255 interact with cellular proteins other than microtubuli, remains open for experimental verification. We went on to define the underlying mechanism of taumediated neurodegeneration by analysis of a large and wide panel of molecular targets and pathways, conform the hypothesis that neurons do not die by a single mechanism. Although the outcome did not yield a single mechanism to be responsible for tau-mediated neuronal cell-death, the indications for attempted cell-cycle reentry were most marked.
involve the microtubular network as a structural and transport scaffold. The alternative explanation, i.e. that wild-type and mutant full-length Tau but not truncated Tau255 interact with cellular proteins other than microtubuli, remains open for experimental verification. We went on to define the underlying mechanism of taumediated neurodegeneration by analysis of a large and wide panel of molecular targets and pathways, conform the hypothesis that neurons do not die by a single mechanism. Although the outcome did not yield a single mechanism to be responsible for tau-mediated neuronal cell-death, the indications for attempted cell-cycle reentry were most marked.
CD is a large molecule and there are contradictory findings in terms of its penetration of the BBB
We add our observation that miRNAmediated deadenylation is evident prior to the onset of measurable translational repression. Taken together, it is thus apparent across different systems that Danshensu miRNAs can stimulate deadenylation without a specific requirement for mRNA translation or a prior translational repression step. Atropine sulfate 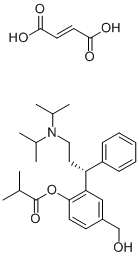 Alternatively, deadenylation may exclusively stimulate mRNA decay. This has been addressed in mammalian and D. melanogaster cells using reporter constructs designed to give rise to mRNAs either carrying the 39 end stem-loop of histone mRNA instead of a tail, or lacking a tail due to the presence of a self-cleaving ribozyme in the 39 UTR. These mRNAs could still be silenced, although at times not as strongly as their polyadenylated counterparts. The common observation is thus that miRNAs can repress translation without a requirement for mRNA deadenylation. It does, however, not necessarily follow that deadenylation has no role in translational repression, as shown by our present findings and previous reports that translational repression is enhanced by the presence of a poly tail. Most importantly, we show here that while blocking let-7-mediated deadenylation does not completely abolish let-7-mediated translational repression, it nevertheless clearly attenuated it. This is a direct demonstration in living cells that mRNA deadenylation can specifically contribute to translational repression by a miRNA. Taken together with the extant literature, our data thus favours a model whereby mRNA deadenylation is a proximal miRNA action that not only promotes mRNA decay but also attenuates the stimulatory role of the poly tail in mRNA translation, by generating an oligo-adenylated form of the mRNA that associates less with cytoplasmic PABP. Indeed, mammalian miRNA target mRNAs are enriched in Ago and GW182 immunoprecipitations, while they are underrepresented in complexes containing PABPC4. The oligo-adenylated mRNA is further silenced by corepressors, which most plausibly interfere with cap/eIF4E function, and then is either stably stored or destroyed by decapping and exonucleolytic degradation. The magnitude of translational repression due to miRNA-mediated deadenylation may depend on the extent to which the poly tail contributed to target mRNA translation before removal; this will vary between mRNAs and with the functional state of the cellular translational machinery. For instance, cells may express variable amounts of several variants of cytoplasmic PABP and PABP activity is know to be regulated by specific inhibitor proteins such as PAIP2. Such differences may explain some of the divergent literature on miRNA-responses to the absence of a poly tail on target mRNA. The balance between decay and storage of target mRNA may further be determined through interplay between the specific sequence context and the given cellular environment. With these provisos, the model explains most reports assessing the role of the poly tail in miRNA-mediated repression. The above scenario clearly does not accommodate postinitiation effects by miRNAs, and several reports have further presented experimental evidence or theoretical arguments to link miRNA-mediated repression to late sub-steps of translation initiation. More work will have to be done to address the possibility that miRNAs affect translation at more than one step. The poly tail also stimulates the 60S subunit joining step during late initiation and it is linked to translation termination through an interaction between PABP and eRF3, providing ways to rationalise a role for deadenylation in several alternative models of translational repression by miRNAs.
Alternatively, deadenylation may exclusively stimulate mRNA decay. This has been addressed in mammalian and D. melanogaster cells using reporter constructs designed to give rise to mRNAs either carrying the 39 end stem-loop of histone mRNA instead of a tail, or lacking a tail due to the presence of a self-cleaving ribozyme in the 39 UTR. These mRNAs could still be silenced, although at times not as strongly as their polyadenylated counterparts. The common observation is thus that miRNAs can repress translation without a requirement for mRNA deadenylation. It does, however, not necessarily follow that deadenylation has no role in translational repression, as shown by our present findings and previous reports that translational repression is enhanced by the presence of a poly tail. Most importantly, we show here that while blocking let-7-mediated deadenylation does not completely abolish let-7-mediated translational repression, it nevertheless clearly attenuated it. This is a direct demonstration in living cells that mRNA deadenylation can specifically contribute to translational repression by a miRNA. Taken together with the extant literature, our data thus favours a model whereby mRNA deadenylation is a proximal miRNA action that not only promotes mRNA decay but also attenuates the stimulatory role of the poly tail in mRNA translation, by generating an oligo-adenylated form of the mRNA that associates less with cytoplasmic PABP. Indeed, mammalian miRNA target mRNAs are enriched in Ago and GW182 immunoprecipitations, while they are underrepresented in complexes containing PABPC4. The oligo-adenylated mRNA is further silenced by corepressors, which most plausibly interfere with cap/eIF4E function, and then is either stably stored or destroyed by decapping and exonucleolytic degradation. The magnitude of translational repression due to miRNA-mediated deadenylation may depend on the extent to which the poly tail contributed to target mRNA translation before removal; this will vary between mRNAs and with the functional state of the cellular translational machinery. For instance, cells may express variable amounts of several variants of cytoplasmic PABP and PABP activity is know to be regulated by specific inhibitor proteins such as PAIP2. Such differences may explain some of the divergent literature on miRNA-responses to the absence of a poly tail on target mRNA. The balance between decay and storage of target mRNA may further be determined through interplay between the specific sequence context and the given cellular environment. With these provisos, the model explains most reports assessing the role of the poly tail in miRNA-mediated repression. The above scenario clearly does not accommodate postinitiation effects by miRNAs, and several reports have further presented experimental evidence or theoretical arguments to link miRNA-mediated repression to late sub-steps of translation initiation. More work will have to be done to address the possibility that miRNAs affect translation at more than one step. The poly tail also stimulates the 60S subunit joining step during late initiation and it is linked to translation termination through an interaction between PABP and eRF3, providing ways to rationalise a role for deadenylation in several alternative models of translational repression by miRNAs.
The potential advantage of data merging was evaluated by means of a quantitative rate of correct classification
The gene signatures were built by a supervised machine learning algorithm like Support Vector Machines or unsupervised classification methods like clustering or statistical method such as Cox regression model and likelihood ratio test. Such gene signatures consist of a list of genes, usually associated with weights that are used to compute a predictive score. Note however that Albaspidin-AA different studies used different performance measures, percentage of concordance in classification to this end. Gene signatures are also Danshensu evaluated in terms of “robustness” and “reproducibility”. Robustness is related to the sample size of the training set from which a gene signature is built and the size of the testing set on which it is validated. A predictor generated from a small training set could have a high prediction accuracy on the training set but may lose generalization power when it is validated on an independent testing set. Moreover, performance estimates obtained with a small testing set have high statistical error, i.e. they come with a large confidence interval. On the other hand, reproducibility means the convergence of results obtained from replicate experiments, possibly carried out in different labs and relying on different technologies. Reproducibility is assessed by cross-data set validation, i.e. the evaluation of a gene signature derived from one data set, with a testing set originating from another study. In this work, we analyzed the potential benefits of merging data sets for prognostic application in breast cancer diagnosis. Contrary to related work, we did not discretize the clinical followup information into good and poor outcome classes, a practice which results in loss of information. Instead, we directly used censored survival data to derive a gene signature that allows for the computation of a risk score from a patients expression profile. The risk score was based on the Cox proportional hazard model, and expected to be inversely related to the time to death or relapse. The basic design of our study is as follows. We used eight breast cancer microarray data sets from eight different studies. Each set had clinical follow-up information in form of censored time to event data, the event being either “overall survival” or “relapse-free survival” or both. The goal was to extract a gene signature from a training set that can be used to predict disease outcome for patients in the testing set. The gene signature 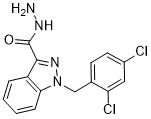 we used consisted of a set of genes plus corresponding Cox coefficients derived by univariate fitting of the expression values to the survival data on the training set. Gene signatures were built from the individual or merged data sets. The accuracy and robustness of prediction were evaluated by 10-fold cross-validation. Reproducibility as defined above was analyzed by training a signature from one or several complete data sets and testing its performance on complete independent validation sets. Data sets were merged in their original numerical representation using two different data integration methods: ComBat and Z-score normalization. Two signature performance measures were computed in each experiment: time dependent Receiver-Operator Characteristic Area Under the Curve and the hazard ratio of the predicted risk scores relative to the survival data in the testing set.
we used consisted of a set of genes plus corresponding Cox coefficients derived by univariate fitting of the expression values to the survival data on the training set. Gene signatures were built from the individual or merged data sets. The accuracy and robustness of prediction were evaluated by 10-fold cross-validation. Reproducibility as defined above was analyzed by training a signature from one or several complete data sets and testing its performance on complete independent validation sets. Data sets were merged in their original numerical representation using two different data integration methods: ComBat and Z-score normalization. Two signature performance measures were computed in each experiment: time dependent Receiver-Operator Characteristic Area Under the Curve and the hazard ratio of the predicted risk scores relative to the survival data in the testing set.
Prediction of flu strains that may cause infection in any flu season cannot accommodate the expected antigenic variability
This illustrates the need for a new vaccine concept that maintains high Mepiroxol protective efficacy regardless of the antigenic variations that arise frequently due to antigenic drift. Chemically and UV inactivated influenza virus preparations rely exclusively on antibody responses for protection and do not induce cytotoxic T cell responses. The Tc cell response to influenza is broadly cross-reactive between virus strains and is important in the recovery from primary infections. We have previously reported that c-Flu preparations can induce crossreactive Tc cell responses. Gamma-irradiation is the preferred method of inactivation of highly infectious agents for biochemical analysis, including Ebola, Marburg and Lassa viruses. It inactivates virus infectivity by generating strand-breaks in the genetic material and has the further advantage, compared with chemical agents, of high penetration into and through biological materials. In contrast to chemical treatment with Atropine sulfate formalin or b-propiolactone, which induces cross-linking of proteins, c-rays have little impact on the antigenic structure and biological integrity of proteins. The Manual on Radiation Sterilization of Medical and Biological Material of the International Atomic Energy Agency indicates that exposure to 0.65 kGy of c-rays causes a total loss of influenza virus infectivity, but disrupting the haemagglutinating activity requires an exposure to higher than 200 kGy. The reduced impact of c-irradiation on the antigenic structure of viral particles is therefore also expected to improve the magnitude and/or quality of humoral immunity elicited in vaccine recipients over that obtained by using present vaccine preparations. We have been investigating the ability of c-Flu preparations to induce heterotypic and cross-protective immunity against avian H5N1 influenza A virus. Our data clearly show that a single intranasal administration of c-A/PR8 protects mice against lethal H5N1 and other heterotypic infections. Human influenza A viruses bind to a2,6-linked sialic acid receptors expressed on epithelial cells of the upper respiratory tract of humans, whereas avian influenza H5N1 viruses bind to a2,3linked receptors expressed predominantly in the lower respiratory tract In general, binding of virus to a2,6-linked receptors is associated with low virulence  but high transmissibility. In contrast, virus binding to a2,3-linked receptors is associated with low transmissibility but high virulence and often lethal influenza virus infections. Therefore, an avian influenza pandemic might be expected to be associated with a mutation in the haemagglutinin molecule that would allow H5N1 virus to bind to a2,6-linked receptors. Subunit vaccines based on currently identified H5 molecules cannot account for the antigenic variants that would result following such mutations and, consequently, neutralizing antibody-mediated protection induced by these vaccines would likely be limited. Tc cell responses to influenza infection, however, are mainly directed against internal viral proteins, which are highly conserved among all influenza A virus strains. Mutations of internal genes are not susceptible to antibodymediated selection and viral escape from Tc cell responses is curtailed by MHC class I polymorphism. Therefore, vaccines inducing Tc cell responses against the highly conserved internal proteins, such as the nucleoprotein, are expected to provide crossprotection against different influenza A virus strains. Our data indicate that the cross-reactive Tc cell responses induced by c-Flu are predominantly directed against the internal proteins, indicated by lysis of NPP-labeled targets.
but high transmissibility. In contrast, virus binding to a2,3-linked receptors is associated with low transmissibility but high virulence and often lethal influenza virus infections. Therefore, an avian influenza pandemic might be expected to be associated with a mutation in the haemagglutinin molecule that would allow H5N1 virus to bind to a2,6-linked receptors. Subunit vaccines based on currently identified H5 molecules cannot account for the antigenic variants that would result following such mutations and, consequently, neutralizing antibody-mediated protection induced by these vaccines would likely be limited. Tc cell responses to influenza infection, however, are mainly directed against internal viral proteins, which are highly conserved among all influenza A virus strains. Mutations of internal genes are not susceptible to antibodymediated selection and viral escape from Tc cell responses is curtailed by MHC class I polymorphism. Therefore, vaccines inducing Tc cell responses against the highly conserved internal proteins, such as the nucleoprotein, are expected to provide crossprotection against different influenza A virus strains. Our data indicate that the cross-reactive Tc cell responses induced by c-Flu are predominantly directed against the internal proteins, indicated by lysis of NPP-labeled targets.