However, various topologies were proposed for Pex11 proteins in different organisms,, and the related Pex11pc was recently reported to dock on the cytosolic site of the peroxisomal membrane. Furthermore, the Tulathromycin B predicted position of the first transmembrane domain within human Pex11pb varies greatly, depending on the in silico search algorithm used, thus resulting e.g. in the designation of Pex11pb as a tail-anchored membrane protein. In the present study, we characterized a newly available Pex11pb antibody directed against an epitope within the putative internal region to determine the topology of Pex11pb. Using differential permeabilization, we confirmed that the epitope recognized by the Pex11pb antibody is only accessible under conditions which permeabilize the peroxisomal membrane. Proteinase K digest of intact peroxisomes and subsequent immunoblotting with the Pex11pb antibody revealed a protease-resistant fragment of approximately 17 kDa, which was degraded upon membrane permeabilization with either Triton X-100 or sonication. The fragment size is consistent with the localization of the first transmembrane domain between aa 90�C110, and the second one between aa 230�C255. These data clearly demonstrate that Pex11pb is an integral membrane protein with two transmembrane spanning domains and N- and C-termini directed towards the 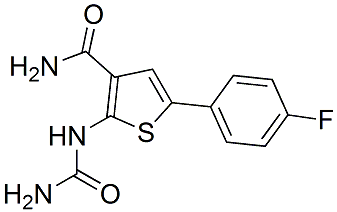 cytosol. The intra-peroxisomal region between the two transmembrane domains is facing the peroxisomal matrix. We cannot rigorously exclude that parts of this region may interact with the matrix site of the peroxisomal membrane, or are partially buried within the membrane. However, deletion of a glycine-rich stretch within the intra-peroxisomal region did not alter the properties of Pex11pb to promote membrane elongation and division of peroxisomes indicating that parts of its internal region are dispensable for these functions. Based on our results on Pex11pb topology, we analyzed putative functional motifs in its sequence and examined their importance for the membrane-shaping properties of Pex11pb. We focused on the cytosolic N-terminal part of the protein which contains three putative amphipathic helices. Pex11pb was the first peroxisomal membrane protein reported to exhibit a special distribution within the peroxisomal membrane as it was found to concentrate in constriction sites on elongated peroxisomes. Furthermore, it preferentially localized to tubular membrane extensions within pre-peroxisomal membrane compartments. A clear preference for tubular membrane structures was also confirmed in this study, as Pex11pb-Myc accumulated in tubular membrane protrusions extending from enlarged peroxisomes which formed under lipidfree culture conditions. Whereas these observations further support a role for Pex11pb in membrane deformation and elongation, the mechanism of its targeting to and retention within these membrane domains remained unclear. In line with this, it has very recently been reported that DHAcontaining phospholipids directly influence homo-oligomerization of Pex11pb, and that incubation of acyl CoA-oxidase deficient fibroblasts with DHA resulted in hyper-oligomerization of Pex11pb giving rise to high molecular mass complexes ranging from 230�C430 kDa. Intriguingly, these findings imply that Pex11pb action on membrane elongation and thus peroxisome division is modulated by phospholipids within the peroxisomal membrane, which in turn are influenced by peroxisomal lipid metabolism such as fatty acid b-oxidation. In previous Butenafine hydrochloride studies, we showed that the addition of polyunsaturated fatty acids promotes the elongatio.
cytosol. The intra-peroxisomal region between the two transmembrane domains is facing the peroxisomal matrix. We cannot rigorously exclude that parts of this region may interact with the matrix site of the peroxisomal membrane, or are partially buried within the membrane. However, deletion of a glycine-rich stretch within the intra-peroxisomal region did not alter the properties of Pex11pb to promote membrane elongation and division of peroxisomes indicating that parts of its internal region are dispensable for these functions. Based on our results on Pex11pb topology, we analyzed putative functional motifs in its sequence and examined their importance for the membrane-shaping properties of Pex11pb. We focused on the cytosolic N-terminal part of the protein which contains three putative amphipathic helices. Pex11pb was the first peroxisomal membrane protein reported to exhibit a special distribution within the peroxisomal membrane as it was found to concentrate in constriction sites on elongated peroxisomes. Furthermore, it preferentially localized to tubular membrane extensions within pre-peroxisomal membrane compartments. A clear preference for tubular membrane structures was also confirmed in this study, as Pex11pb-Myc accumulated in tubular membrane protrusions extending from enlarged peroxisomes which formed under lipidfree culture conditions. Whereas these observations further support a role for Pex11pb in membrane deformation and elongation, the mechanism of its targeting to and retention within these membrane domains remained unclear. In line with this, it has very recently been reported that DHAcontaining phospholipids directly influence homo-oligomerization of Pex11pb, and that incubation of acyl CoA-oxidase deficient fibroblasts with DHA resulted in hyper-oligomerization of Pex11pb giving rise to high molecular mass complexes ranging from 230�C430 kDa. Intriguingly, these findings imply that Pex11pb action on membrane elongation and thus peroxisome division is modulated by phospholipids within the peroxisomal membrane, which in turn are influenced by peroxisomal lipid metabolism such as fatty acid b-oxidation. In previous Butenafine hydrochloride studies, we showed that the addition of polyunsaturated fatty acids promotes the elongatio.
Monthly Archives: June 2019
Modes of action of the DNA methylation and Polycomb systems in contributing to cell identity
Dnmt1 null ESCs differentiated as embryoid bodies express high levels of trophoblast markers and, under appropriate conditions, can be differentiated into trophoblast derivatives. Similarly, it has been reported that TKO ESCs exhibit a growth defect and increased apoptosis upon EB differentiation and that cells from TKO nuclear transfer embryos aggregated with wt embryos mostly contribute to extraembryonic tissues, in addition to. However, in the latter study a few TKO cells were detected in the embryo proper till an early postgastrulation stage, but their 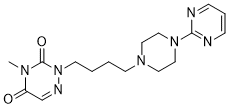 identity was not defined. Thus, it is not clear to what extent globally hypomethylated cells are able to commit to and progress along definitive embryonic lineages. The role of DNA methylation in controlling transcription programs during mammalian development and lineage specification has been mainly inferred from genomic methylcytosine profiles in a limited selection of cell types and developmental stages, while little information is available about how DNA methylation and Dnmts actually affect transcription during differentiation. In Benzethonium Chloride particular, expression data from differentiated progeny of globally hypomethylated ESCs lacking specific Dnmts are very scarce. This is likely in relation to reports of limited survival or proliferation of Dnmt1 null and TKO cells upon differentiation. Consistent with this previous work, we found that the average size of Dnmt1 null and especially TKO EBs is reduced as compared to that of wt EBs. However, we could maintain cultures of these mutant EBs for at least 24 days. EB formation is an undirected differentiation model supporting the specification of a broad range of cell fates and thus commonly used to asses developmental potential. Using this system we found that Dnmt1 null and TKO EBs exhibit residual transcription of pluripotency master regulators Oct4 and Nanog even after 16 days of culture. However, FACS analysis showed that by day 8 Oct4 protein is uniformly downregulated in all the cells of these mutant EBs to the same basal levels as in wt EBs, suggesting homogeneous exit from the ESC state. This is supported by the relatively high concordance of genome-wide transcription changes in mutant relative to wt EBs after 4 days of differentiation, including downregulation of additional genes associated with pluripotent stem cell states and upregulation of factors related to differentiated lineages. However, by day 16 gene expression profiles in TKO EBs reveal a high degree of divergence from those in wt EBs, while expression changes in Dnmt1 null EBs are still concordant with one third of all changes in wt EBs. On the one hand this represents a previously unappreciated progression of transcription programs in differentiated Dnmt1 null cells, on the other hand it reflects substantially impaired developmental Gomisin-D potential in TKO as well as Dnmt1 null cells. In particular, ESCs lacking both PRC1 and 2 show twice the number of derepressed genes as ESCs lacking either complex, where little derepression of ERVs is observed, leading to the proposal that repetitive sequences serve as a platform for gene silencing by PRC complexes. This is reminiscent of the higher number of disregulated genes in TKO versus Dnmt1 null EBs and of substantial residual methylation of repetitive sequences in Dnmt1 null cells. It is therefore tempting to speculate that methylation of repetitive sequences may also serve as a platform for gene silencing upon differentiation.
identity was not defined. Thus, it is not clear to what extent globally hypomethylated cells are able to commit to and progress along definitive embryonic lineages. The role of DNA methylation in controlling transcription programs during mammalian development and lineage specification has been mainly inferred from genomic methylcytosine profiles in a limited selection of cell types and developmental stages, while little information is available about how DNA methylation and Dnmts actually affect transcription during differentiation. In Benzethonium Chloride particular, expression data from differentiated progeny of globally hypomethylated ESCs lacking specific Dnmts are very scarce. This is likely in relation to reports of limited survival or proliferation of Dnmt1 null and TKO cells upon differentiation. Consistent with this previous work, we found that the average size of Dnmt1 null and especially TKO EBs is reduced as compared to that of wt EBs. However, we could maintain cultures of these mutant EBs for at least 24 days. EB formation is an undirected differentiation model supporting the specification of a broad range of cell fates and thus commonly used to asses developmental potential. Using this system we found that Dnmt1 null and TKO EBs exhibit residual transcription of pluripotency master regulators Oct4 and Nanog even after 16 days of culture. However, FACS analysis showed that by day 8 Oct4 protein is uniformly downregulated in all the cells of these mutant EBs to the same basal levels as in wt EBs, suggesting homogeneous exit from the ESC state. This is supported by the relatively high concordance of genome-wide transcription changes in mutant relative to wt EBs after 4 days of differentiation, including downregulation of additional genes associated with pluripotent stem cell states and upregulation of factors related to differentiated lineages. However, by day 16 gene expression profiles in TKO EBs reveal a high degree of divergence from those in wt EBs, while expression changes in Dnmt1 null EBs are still concordant with one third of all changes in wt EBs. On the one hand this represents a previously unappreciated progression of transcription programs in differentiated Dnmt1 null cells, on the other hand it reflects substantially impaired developmental Gomisin-D potential in TKO as well as Dnmt1 null cells. In particular, ESCs lacking both PRC1 and 2 show twice the number of derepressed genes as ESCs lacking either complex, where little derepression of ERVs is observed, leading to the proposal that repetitive sequences serve as a platform for gene silencing by PRC complexes. This is reminiscent of the higher number of disregulated genes in TKO versus Dnmt1 null EBs and of substantial residual methylation of repetitive sequences in Dnmt1 null cells. It is therefore tempting to speculate that methylation of repetitive sequences may also serve as a platform for gene silencing upon differentiation.
In some instances this enhancement varies depending on the target microorganism but in other cases
This enhancement is consistent across a wide range of targets. The fact that this enhancement is apparent against Gram positive and Gram negative targets is particularly novel. Further efforts will focus on determining the mechanistic basis for these enhancements and an Mepiroxol assessment of how well these peptides perform in food and in vivo. The mechanism by which the developing crop influences return bloom and yield the following year is not fully understood. Two hypotheses have been suggested. The “nutritional” hypothesis holds that return bloom and yield are proportional to tree carbohydrate status. Lack of carbohydrate in the ON year Catharanthine sulfate directly or indirectly reduces flowering the following year. Support for this hypothesis has been provided by showing positive correlations between carbohydrate levels and AB status, whereas others have shown no consistent relationship between tree carbohydrate status and floral intensity at return bloom. The “hormonal” hypothesis proposes that developing fruit produce an inhibitor that directly or indirectly reduces flowering in the spring following the ON crop. Although a number of studies have shown correlations between abscisic acid or indole-3-acetic acid and AB status, no direct evidence has been provided for their involvement in the return bloom. Gibberellin is well-known inhibitor of flowering in citrus; thus, fruitproduced GA has been presumed to be involved in AB. Despite these findings, the roles of carbohydrates and hormones in AB remain unclear and more research is needed to identify factors affecting floral intensity following ON and OFF years. Genetic analysis of AB in apple identified a few QTLs associated with AB, and suggested that hormone-related genes are likely to play a role in the phenomenon. The floral induction period in citrus starts in mid-November and lasts until approximately the end of December to mid-January. Following induction, the bud enters a short resting period, after which the shoot apical meristem differentiates into a floral bud. In parallel to the floral shoot flush, there is a flush of vegetative shoot growth, which continues through June. A second flush of vegetative shoot growth starts in July, and third flush starts in October. Usually, next year flowering occurs mostly on the spring vegetative flush. Flowering in citrus is induced by low temperature, while day length has a relatively minor effect. There is extensive cross-talk between autonomous and vernalization flowering pathways and ample evidence that genes associated with flowering regulation are highly conserved across species. Fruit presence inhibits return flowering. However, it is not clear at which stage the fruit exerts its inhibitory effect: at flowering induction, transition of the shoot apical meristem to floral meristem, or subsequent stages of floral development and bud break. Moreover, the nature of the signal and the organ or tissue from which it originates, be it the fruit itself or the leaf which senses fruit presence, are not known. Regardless of the source tissue for the AB signal, it must be received, directly or indirectly, at the bud, and more specifically, at the apical meristem which has to “decide” whether to develop into an inflorescence or remain a vegetative meristem. Therefore, following perception of the signal, the bud must undergo a series of events which depend on fruit load. In the current work, we analyzed changes in global gene expression during bud development in ON and OFF trees, to identify metabolic and controlling pathways that play a role in 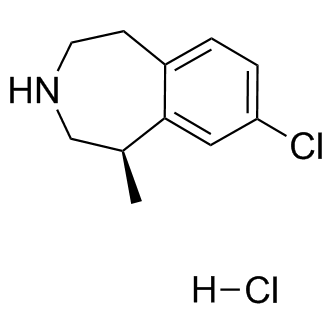 bud fate. To determine the earliest time point for the transcriptome analysis, we first analyzed changes in bud morphology during its development, and changes in the expression of key flowering control genes.
bud fate. To determine the earliest time point for the transcriptome analysis, we first analyzed changes in bud morphology during its development, and changes in the expression of key flowering control genes.
Despite these advances a detailed understanding of the role of hormones involve a series of low-affinity
Non-covalent and reversible interactions with FG domains from multiple Nups and diffusion through the NPC, whereas interaction with Nup153 might be critical for mediating irreversible and directional exit from the NPC. Interaction with FG-repeats may be mediated indirectly by nuclear transport receptors, such as Mechlorethamine hydrochloride importin 7,, or transportin 3. After exit from the nuclear basket, both Nup98 and Nup153, which shuttle on and off the NPC and have been shown to interact with chromatin,,, might accompany the PIC to its integration site. In particular, Nup98 has been found to localise to the nucleoplasm and participate in chromosomal remodelling and regulation of gene expression. It will be interesting to determine whether Nup98 and/or Nup153 are hijacked by HIV-1 for transport to euchromatin and contribute to specific site selection in expressed genes, since previous work has shown that HIV-1 integrates preferentially within actively transcribed genes. Surprisingly, Nup98 depletion affected HIV-1 and MLV infection equally, but the reduction of MLV infectivity could merely be due to the slight accumulation of cells in G1/S phase that we observed since MLV enters the nucleus during metaphase. Our work demonstrates that a key to the ability of HIV-1 to replicate in non-dividing cells is its capacity to use NPC components for its active transport across the nuclear pore, thus underlining the evolutionary adaptability of HIV-1 to exploit host mechanisms to achieve active nuclear import. Our study suggests a new appealing role for the NPC in HIV-1 infection proposing that the viral nuclear entry step may be important not only for actual translocation, but also for correct subsequent integration as a result of the physical interaction that exists between nuclear pore baskets and the chromatin. The study of the physical and functional interactions between HIV-1 and the NPC not only contributes to our understanding of how other viruses manipulate the nuclear pore but also strengthen our comprehension of lentiviral vectors used for gene transfer protocols, whose active nuclear import is similar to 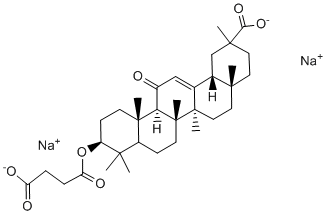 that of HIV-1. From a seed no bigger than that of a cucumber, California’s coastal redwood tree can grow to a height of more than 350 feet. At the same time, aquatic watermeal plants are so small that they resemble specks of cornmeal. The nature of the mechanisms controlling the size and shape of organs is an important but still unanswered question of developmental biology. Two factors determine the size of mature organs: cell number and cell size. A current model for metazoans proposes that cell number and cell size are controlled by distinct proliferation and growth signals that negatively affect each other; when cellular size is increased, cell count is reduced and vice versa. This regulation ensures that induced alterations in cell proliferation are compensated for by changes in cell size, resulting in little net Benzethonium Chloride change in the final organ size. The compensation phenomenon has also been observed in plants �C; however, it is not clear whether the model proposed for metazoans can be directly applied to plants;. First of all, the majority of cell elongation occurs after termination of cell division and, therefore, the actions of proliferation factors and cell expansion factors are separated in time. In addition, some signals, such as auxin, regulate both proliferation and elongation of cells . To understand regulation of organ size and the molecular mechanism of compensation it is essential to know the signaling pathways that control and coordinate cell proliferation and cell elongation. Plant hormones are the most obvious candidates for that role, with cytokinins being implicated in regulation of cell proliferation; and gibberellins, brassinosteroids, and auxin regulating both cell proliferation and cell elongation.
that of HIV-1. From a seed no bigger than that of a cucumber, California’s coastal redwood tree can grow to a height of more than 350 feet. At the same time, aquatic watermeal plants are so small that they resemble specks of cornmeal. The nature of the mechanisms controlling the size and shape of organs is an important but still unanswered question of developmental biology. Two factors determine the size of mature organs: cell number and cell size. A current model for metazoans proposes that cell number and cell size are controlled by distinct proliferation and growth signals that negatively affect each other; when cellular size is increased, cell count is reduced and vice versa. This regulation ensures that induced alterations in cell proliferation are compensated for by changes in cell size, resulting in little net Benzethonium Chloride change in the final organ size. The compensation phenomenon has also been observed in plants �C; however, it is not clear whether the model proposed for metazoans can be directly applied to plants;. First of all, the majority of cell elongation occurs after termination of cell division and, therefore, the actions of proliferation factors and cell expansion factors are separated in time. In addition, some signals, such as auxin, regulate both proliferation and elongation of cells . To understand regulation of organ size and the molecular mechanism of compensation it is essential to know the signaling pathways that control and coordinate cell proliferation and cell elongation. Plant hormones are the most obvious candidates for that role, with cytokinins being implicated in regulation of cell proliferation; and gibberellins, brassinosteroids, and auxin regulating both cell proliferation and cell elongation.
Be causative of disease and may require additional factors for a phenotypic manifestation
Gene-expression analysis was carried out at a few key time points in the buds, which receive the ‘AB signal’, and in leaves and stems, which might play a role in generating and transporting the signal. Our resequencing study reported here and those reported elsewhere Cinoxacin indicate that non-synonymous mutations of CITED2 can be observed in patients with congenital heart disease, and that these mutations tend to cluster in the SRJ domain. Cellbased assays investigating some of these variants have indicated that they can affect HIF1A-repression and/or TFAP2-coactivation functions of CITED2. The SRJ domain, although highly conserved in placental mammals, is substantially abbreviated in marsupials and in monotremes, and is absent in other vertebrates. Thus, the region may have appeared relatively recently in evolutionary terms and may conceivably be of relevance to differences in cardiac development and structure between placental mammals and other vertebrates. Structurally, the SRJ region is predicted to be disordered and potentially functions as a flexible linker. The T166 residue within this domain is predicted to be a target of proline directed kinases, and our studies indicated that the T166 residue can be phosphorylated by MAPK1, and that activation of MAPK1 promoted co-activation function. Moreover cell-based studies indicated that the T166N mutation had a deleterious effect on TFAP2 co-activation function, and on the ability of CITED2 to promote ES cell proliferation in the absence of LIF. Surprisingly, our in vivo study of mice carrying the T166N 3,4,5-Trimethoxyphenylacetic acid variant and the deletion of the SRJ domain indicate that in mice, under normal laboratory conditions, neither the complete deletion of the SRJ domain nor the introduction of a variant which could potentially lead to the loss of a phosphorylation site of CITED2 are detrimental to its function. In a single Cited2 2/MRG1 embryo out of 75 that we studied, we observed an ectopia cordis phenotype. Since ectopia cordis is not a phenotype which has previously been associated with the loss of Cited2, and this embryo has normal adrenal glands, it is most unlikely that it is a consequence of the loss of the SRJ domain. Animals of this genotype are also viable and fertile. However, in vitro data indicate that the T166N mutation can have functional significance, and previous studies have indicated that other SRJ mutations also affect its function. It is possible that the partial impairment of CITED2 function revealed by in vitro experiments is insufficient to affect development. Taken together, the results obtained from mouse studies indicate that the SRJ domain is dispensable during mouse cardiac development and for viability and fertility. On the other hand, three independent human studies show that non-synonymous mutations, predominantly clustering in the SRJ domain, are mainly observed in patients with CHD and not in controls suggesting that  this region is important for normal cardiac development. How can we explain these divergent observations? One possibility is that the SRJ is indeed dispensable for mammalian heart development, and that the observations from patients may be misleading. Supporting this idea, there is considerable lack of conservation in this domain between placental and non-placental mammals. Furthermore, the mutations in the SRJ do not significantly affect the disordered nature of the domain, indicating that it may be able to accommodate mutations without adversely affecting the overall structure and function of the protein. Moreover, in no case has it been shown that the mutation has either arisen de novo or been transmitted from an affected parent. Another possibility that partially reconciles the mouse and human observations is that variants found clustered in the SRJ in CHD patients may not, by themselves.
this region is important for normal cardiac development. How can we explain these divergent observations? One possibility is that the SRJ is indeed dispensable for mammalian heart development, and that the observations from patients may be misleading. Supporting this idea, there is considerable lack of conservation in this domain between placental and non-placental mammals. Furthermore, the mutations in the SRJ do not significantly affect the disordered nature of the domain, indicating that it may be able to accommodate mutations without adversely affecting the overall structure and function of the protein. Moreover, in no case has it been shown that the mutation has either arisen de novo or been transmitted from an affected parent. Another possibility that partially reconciles the mouse and human observations is that variants found clustered in the SRJ in CHD patients may not, by themselves.