The prediction of drug-target interactions can be directly applied to completing the genome annotations, investigating the drug Dexrazoxane hydrochloride specificity and promiscuity, and finding the targets for diseases. However, the overall pattern of the interaction interface between the chemical space and biological systems is too large and complex to be captured. For simplicity, the enzyme which has been extensively studied as a class of important drug target family was selected here to illustrate our models’ applications of predicting the potential drug-target interactions. A total of 175 enzymes in the DrugBank database were matched with each of 6511 drugs to conduct the comprehensive drug-target interaction prediction using the general RF Model I. Interestingly, this demonstrates that only a few families of enzymes and drugs account for the top scoring interactions, which is completely supported by a previous model LOUREIRIN-B established based on the bipartite graph learning algorithm. Meanwhile, the results also demonstrate that our RF approach offers better predictions for proteins/chemicals that have been extensively studied and for which many ligands/ receptors are known. These results further imply that our proposed methods are able to learn, i.e., the more information is provided, the better the prediction. Here, we take the top 10 scoring novel interactions as examples to illustrate the above findings. As shown in Table 3, compound Bromfenac is predicted to act on Prostaglandin G/H synthase 1 with a score of 0.992. Actually, this interaction has been confirmed and was annotated in DrugBank database. For Asparagine synthetase, a new ligand Indomethacin is predicted to bind to it with the score of 0.984, which might be hinted by an indirect experiment in which the Asparagine synthetase expression level was indeed upregulated by this compound. Additionally, Oxyphenbutazone, as a well-known nonsteroidal anti-inflammatory agent, binds to the cyclooxygenase Prostaglandin G/H synthase 1 and Prostaglandin G/H synthase 2 with 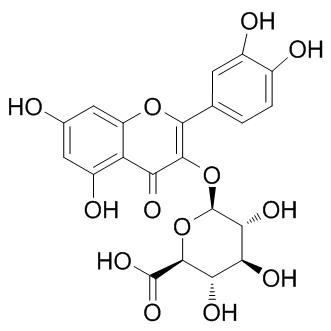 the same binding scores of 0.982. This is supported by the fact that the nonsteroidal anti-inflammatory drugs could produce therapeutic activities through the inhibition of cyclooxygenase. With no exception, for all the remaining interactions, the predicted ligands of certain receptors are invariably similar in structure with those confirmed ligands as mentioned above. All these outcomes enhance the strength of our proposed methods for realistic drugtarget interaction prediction application. Subsequently, a comprehensive network describing the drugtarget interactions was constructed. In order to make it clear and simple, the top 500 scoring drug-target interactions were used to generate a bipartite graph of drug-target interactions for illustrating the complex relationships between drugs and enzymes, in which a compound and a protein are connected to each other if the protein is a predicted target of the compound. Figure 7 shows a global view of this network with color and shape-coded nodes. To explore the topological and global properties of this drug-target network, the centralization, heterogeneity and node degree distribution were analyzed. The centralization and heterogeneity analysis shows the network centralization and heterogeneity degrees are 0.463 and 3.661, respectively, indicating that a few nodes are more central that the other ones in this net, i.e., the drugtarget space is biased toward certain compounds and proteins.
the same binding scores of 0.982. This is supported by the fact that the nonsteroidal anti-inflammatory drugs could produce therapeutic activities through the inhibition of cyclooxygenase. With no exception, for all the remaining interactions, the predicted ligands of certain receptors are invariably similar in structure with those confirmed ligands as mentioned above. All these outcomes enhance the strength of our proposed methods for realistic drugtarget interaction prediction application. Subsequently, a comprehensive network describing the drugtarget interactions was constructed. In order to make it clear and simple, the top 500 scoring drug-target interactions were used to generate a bipartite graph of drug-target interactions for illustrating the complex relationships between drugs and enzymes, in which a compound and a protein are connected to each other if the protein is a predicted target of the compound. Figure 7 shows a global view of this network with color and shape-coded nodes. To explore the topological and global properties of this drug-target network, the centralization, heterogeneity and node degree distribution were analyzed. The centralization and heterogeneity analysis shows the network centralization and heterogeneity degrees are 0.463 and 3.661, respectively, indicating that a few nodes are more central that the other ones in this net, i.e., the drugtarget space is biased toward certain compounds and proteins.
Low degrees with only a small proportion of components interacting with the multiple partners
Leave a reply